Generated project files¶
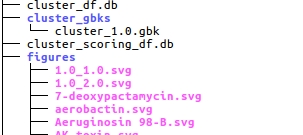
The ‘.db’ files are MySQL dumps of the DataFrames generated for the gene cluster. The can be observed with various programs such as SQLlite).
- cluster_df.db contains all information about the detected gene clusters.
- cluster_scoring_df.db contains all information about the natural product screening.
- gbk_df.db contains all information about the detected domains.
- meta_df.db contains all meta information such as file name and description.
The folder cluster_gbks contains annotated GBK files for each cluster.
The figures folder contains images of the compounds from the natural product screening. Each figure corresponds to one compound listed in cluster_scoring_df.db.
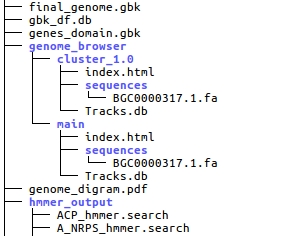
The genes_genome.gbk is a minimal annotation of the genome (only includes predicted domains). The final_genome.gbk is a complete annotation of the genome (included domains, modules, cluster …).
The genome browser folder allows to run the genome browser locally. This only works when a server is simulated. This can be done with python: Just run:
python -m http.server 8000
in the folder. And then open http://127.0.0.1:8000/ in any browser.
An additional “old” genome_diagram as PDF is also included.
The hmmer_output folder contains all hmmer results for each domain.
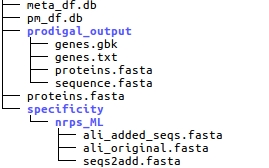
The prodigal_output folder contains all the results of the Prodigal gene prediction.
The specificity folder contains the alignments used for the specificity prediction.